Agenda
📚 Multi-xPU thermal porous convection 3D
💻 Automatic documentation an CI
🚧 Project:
Multi-xPU thermal porous convection 3D
Automatic documentation and CI
Projects
Create a multi-xPU version of the 3D thermal porous convection xPU code
Combine ImplicitGlobalGrid.jl and ParallelStencil.jl
Finalise the documentation of your project
Automatic documentation and CI
ImplicitGlobalGrid.jl (continued)In previous Lecture 8, we introduced ImplicitGlobalGrid.jl, which renders distributed parallelisation with GPU and CPU for HPC a very simple task.
Also, ImplicitGlobalGrid.jl elegantly combines with ParallelStencil.jl to, e.g., hide communication behind computation.
Let's have a rapid tour of ImplicitGlobalGrid.jl's' documentation before using it to turn the 3D thermal porous diffusion solver into a multi-xPU solver.
Let's step through the following content:
Create a multi-xPU version of your thermal porous convection 3D xPU code you finalised in lecture 7
Keep it xPU compatible using ParallelStencil.jl
Deploy it on multiple xPUs using ImplicitGlobalGrid.jl
👉 You'll find a version of the PorousConvection_3D_xpu.jl code in the solutions folder on Moodle after exercises deadline if needed to get you started.
Only a few changes are required to enable multi-xPU support, namely:
Copy your working PorousConvection_3D_xpu.jl code developed for the exercises in Lecture 7 and rename it PorousConvection_3D_multixpu.jl.
Add at the beginning of the code
using ImplicitGlobalGrid
import MPIFurther, add global maximum computation using MPI reduction function to be used instead of maximum()
max_g(A) = (max_l = maximum(A); MPI.Allreduce(max_l, MPI.MAX, MPI.COMM_WORLD))In the # numerics section, initialise the global grid right after defining nx,ny,nz and use now global grid nx_g(),ny_g() and nz_g() for defining maxiter and ncheck, as well as in any other places when needed.
nx,ny = 2 * (nz + 1) - 1, nz
me, dims = init_global_grid(nx, ny, nz) # init global grid and more
b_width = (8, 8, 4) # for comm / comp overlapModify the temperature initialisation using ImplicitGlobalGrid's global coordinate helpers (x_g, etc...), including one internal boundary condition update (update halo):
T = @zeros(nx, ny, nz)
T .= Data.Array([ΔT * exp(-(x_g(ix, dx, T) + dx / 2 - lx / 2)^2
-(y_g(iy, dy, T) + dy / 2 - ly / 2)^2
-(z_g(iz, dz, T) + dz / 2 - lz / 2)^2) for ix = 1:size(T, 1), iy = 1:size(T, 2), iz = 1:size(T, 3)])
T[:, :, 1 ] .= ΔT / 2
T[:, :, end] .= -ΔT / 2
update_halo!(T)
T_old = copy(T)Prepare for visualisation, making sure only me==0 creates the output directory. Also, prepare an array for storing inner points only (no halo) T_inn as well as global array to gather subdomains T_v
if do_viz
ENV["GKSwstype"]="nul"
if (me==0) if isdir("viz3Dmpi_out")==false mkdir("viz3Dmpi_out") end; loadpath="viz3Dmpi_out/"; anim=Animation(loadpath,String[]); println("Animation directory: $(anim.dir)") end
nx_v, ny_v, nz_v = (nx - 2) * dims[1], (ny - 2) * dims[2], (nz - 2) * dims[3]
(nx_v * ny_v * nz_v * sizeof(Data.Number) > 0.8 * Sys.free_memory()) && error("Not enough memory for visualization.")
T_v = zeros(nx_v, ny_v, nz_v) # global array for visu
T_inn = zeros(nx - 2, ny - 2, nz - 2) # no halo local array for visu
xi_g, zi_g = LinRange(-lx / 2 + dx + dx / 2, lx / 2 - dx - dx / 2, nx_v), LinRange(-lz + dz + dz / 2, -dz - dz / 2, nz_v) # inner points only
iframe = 0
endMoving to the time loop, add halo update function update_halo! after the kernel that computes the fluid fluxes. You can additionally wrap it in the @hide_communication block to enable communication/computation overlap (using b_width defined above)
@hide_communication b_width begin
@parallel compute_Dflux!(qDx, qDy, qDz, Pf, T, k_ηf, _dx, _dy, _dz, αρg, _1_θ_dτ_D)
update_halo!(qDx, qDy, qDz)
endApply a similar step to the temperature update, where you can also include boundary condition computation as following (⚠️ no other construct is currently allowed)
@hide_communication b_width begin
@parallel update_T!(T, qTx, qTy, qTz, dTdt, _dx, _dy, _dz, _1_dt_β_dτ_T)
@parallel (1:size(T, 2), 1:size(T, 3)) bc_x!(T)
@parallel (1:size(T, 1), 1:size(T, 3)) bc_y!(T)
update_halo!(T)
endUse now the max_g function instead of maximum to collect the global maximum among all local arrays spanning all MPI processes. Use it in the timestep dt definition and in the error calculation (instead of maximum).
# time step
dt = if it == 1
0.1 * min(dx, dy, dz) / (αρg * ΔT * k_ηf)
else
min(5.0 * min(dx, dy, dz) / (αρg * ΔT * k_ηf), ϕ * min(dx / max_g(abs.(qDx)), dy / max_g(abs.(qDy)), dz / max_g(abs.(qDz))) / 3.1)
endMake sure all printing statements are only executed by me==0 in order to avoid each MPI process to print to screen, and use nx_g() instead of local nx in the printed statements when assessing the iteration per number of grid points.
Update the visualisation and output saving part
# visualisation
if do_viz && (it % nvis == 0)
T_inn .= Array(T)[2:end-1, 2:end-1, 2:end-1]; gather!(T_inn, T_v)
if me == 0
p1 = heatmap(xi_g, zi_g, T_v[:, ceil(Int, ny_g() / 2), :]'; xlims=(xi_g[1], xi_g[end]), ylims=(zi_g[1], zi_g[end]), aspect_ratio=1, c=:turbo)
# display(p1)
png(p1, @sprintf("viz3Dmpi_out/%04d.png", iframe += 1))
save_array(@sprintf("viz3Dmpi_out/out_T_%04d", iframe), convert.(Float32, T_v))
end
endFinalise the global grid before returning from the main function
finalize_global_grid()
returnIf you made it up to here, you should now be able to launch your PorousConvection_3D_multixpu.jl code on multiple GPUs. Let's give it a try 🔥
Make sure to have set following parameters:
lx,ly,lz = 40.0, 20.0, 20.0
Ra = 1000
nz = 63
nx,ny = 2 * (nz + 1) - 1, nz
b_width = (8, 8, 4) # for comm / comp overlap
nt = 500
nvis = 50Then, launch the script on Piz Daint on 8 GPU nodes upon adapting the the runme_mpi_daint.sh or sbatch sbatch_mpi_daint.sh scripts (see here) 🚀
The final 2D slice (at ny_g()/2) produced should look as following and the code takes about 25min to run:
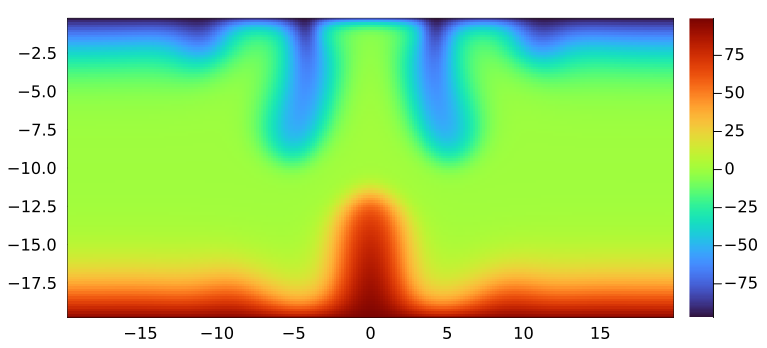
Running the code at higher resolution (508x252x252 grid points) and for 6000 timesteps produces the following result
This lecture we will learn:
documentation vs code-comments
why to write documentation
GitHub tools:
rendering of markdown files
gh-pages
some Julia tools:
Why should I write code comments?
"Code Tells You How, Comments Tell You Why"
code should be made understandable by itself, as much as possible
comments then should be to tell the "why" you're doing something
but I do a lot of structuring comments as well
math-y variables tend to be short and need a comment as well
Why should I write documentation?
documentation should give a bigger overview of what your code does
at the function-level (doc-strings)
at the package-level (README, full-fledged documentation)
to let other people and your future self (probably most importantly) understand what your code is about
Worse than no documentation/code comments is documentation which is outdated.
I find the best way to keep documentation up to date is:
have documentation visible to you, e.g. GitHub README
document what you need yourself
use examples and run them as part of CI (doc-tests, example-scripts)
A Julia doc-string (Julia manual):
is just a string before the object (no blank-line inbetween); interpreted as markdown-string
can be attached to most things (functions, variables, modules, macros, types)
can be queried with ?
"Typical size of a beer crate"
const BEERBOX = 12?BEERBOXOne can add examples to doc-strings (they can even be part of testing: doc-tests).
(Run it in the REPL and copy paste to the docstring.)
"""
transform(r, θ)
Transform polar `(r,θ)` to cartesian coordinates `(x,y)`.
# Example
```jldoctest
julia> transform(4.5, pi/5)
(3.6405764746872635, 2.6450336353161292)
```
"""
transform(r, θ) = (r*cos(θ), r*sin(θ))?transformThe easiest way to write long-form documentation is to just use GitHub's markdown rendering.
A nice example is this short course by Ludovic (incidentally about solving PDEs on GPUs 🙂).
images are rendered
in-page links are easy, e.g. [_back to workshop material_](#workshop-material)
top-left has a burger-menu for page navigation
can be edited within the web-page (pencil-icon)
👉 this is a good and low-overhead way to produce pretty nice documentation
There are several tools which render .jl files (with special formatting) into markdown files. These files can then be added to GitHub and will be rendered there.
we're using Literate.jl
format is described here
files stay valid Julia scripts, i.e. they can be executed without Literate.jl
Example
input julia-code in: course-101-0250-00-L8Documentation.jl: scripts/car_travels.jl
output markdown in: course-101-0250-00-L8Documentation.jl: scripts/car_travels.md created with:
Literate.markdown("car_travels.jl", directory_of_this_file, execute=true, documenter=false, credit=false)But this is not automatic! Manual steps: run Literate, add files, commit and push...
or use GitHub Actions...
Demonstrated in the repo course-101-0250-00-L8Documentation.jl
name: Run Literate.jl
# adapted from https://lannonbr.com/blog/2019-12-09-git-commit-in-actions
on: push
jobs:
lit:
runs-on: ubuntu-latest
steps:
# Checkout the branch
- uses: actions/checkout@v2
- uses: julia-actions/setup-julia@v1
with:
version: '1.9'
arch: x64
- uses: julia-actions/cache@v1
- uses: julia-actions/julia-buildpkg@latest
- name: run Literate
run: QT_QPA_PLATFORM=offscreen julia --color=yes --project -e 'cd("scripts"); include("literate-script.jl")'
- name: setup git config
run: |
# setup the username and email. I tend to use 'GitHub Actions Bot' with no email by default
git config user.name "GitHub Actions Bot"
git config user.email "<>"
- name: commit
run: |
# Stage the file, commit and push
git add scripts/md/*
git commit -m "Commit markdown files fom Literate"
git push origin masterIf you want to have full-blown documentation, including, e.g., automatic API documentation generation, versioning, then use Documenter.jl.
Examples:
Notes:
it's geared towards Julia-packages, less for a bunch-of-scripts as in our lecture
Documenter.jl also integrates with Literate.jl.
for more free-form websites, use https://github.com/tlienart/Franklin.jl (as the course website does)
if you want to use it, it's easiest to generate your package with PkgTemplates.jl which will generate the Documenter-setup for you.
we don't use it in this course
PorousConvection subfolder in your private GitHub repo. The git commit hash (or SHA) of the final push needs to be uploaded on Moodle (more).homework-7 branch, create a new git branch named homework-9 in order to build upon work performed for homework 7.👉 See Logistics for submission details.
The goal of this exercise is to:
Further familiarise with distributed computing
Combine ImplicitGlobalGrid.jl and ParallelStencil.jl
Learn about GPU MPI on the way
In this exercise, you will:
Create a multi-xPU version of the 3D thermal porous convection code from lecture 7
Keep it xPU compatible using ParallelStencil.jl
Deploy it on multiple xPUs using ImplicitGlobalGrid.jl
👉 You'll find a version of the PorousConvection_3D_xpu.jl code on Moodle after exercises deadline if needed to get you started.
Copy the PorousConvection_3D_xpu.jl code from exercises in Lecture 7 and rename it PorousConvection_3D_multixpu.jl.
Refer to the steps outlined in the Multi-xPU 3D thermal porous convection section from Lecture 9 to implement the changes needed to port the 3D single xPU code (from Lecture 7) to multi-xPU.
Upon completion, verify the script converges and produces expected output for following parameters:
lx,ly,lz = 40.0, 20.0, 20.0
Ra = 1000
nz = 63
nx,ny = 2 * (nz + 1) - 1, nz
b_width = (8, 8, 4) # for comm / comp overlap
nt = 500
nvis = 50Use 8 GPUs on Piz Daint adapting the runme_mpi_daint.sh or sbatch sbatch_mpi_daint.sh scripts (see here) to use CUDA-aware MPI 🚀
The final 2D slice (at ny_g()/2) produced should look similar as the figure depicted in Lecture 9.
Now that you made sure the code runs as expected, launch PorousConvection_3D_multixpu.jl for 2000 steps on 8 GPUs at higher resolution (global grid of 508x252x252) setting:
nz = 127
nx,ny = 2 * (nz + 1) - 1, nz
nt = 2000
nvis = 100and keeping other parameters unchanged.
Use sbtach command to launch a non-interactive job which may take about 5h30-6h to execute.
Produce a figure or animation showing the final stage of temperature distribution in 3D and add it to a new section titled ## Porous convection 3D MPI in the PorousConvection project subfolder's README.md. You can use the Makie visualisation helper script from Lecture 7 for this purpose (making sure to adapt the resolution and other input params if needed).
👉 See Logistics for submission details.
The goal of this exercise is to:
write some documentation
using doc-strings
using Literate.jl
One task you've already done, namely to update the README.md of this set of exercises!
Tasks:
Add doc-string to the functions of following scripts:
PorousConvection_3D_xpu.jl
PorousConvection_3D_multixpu.jl
Add to the PorousConvection folder a Literate.jl script called bin_io_script.jl that contains and documents following save_array and load_array functions you may have used in your 3D script
"""
Some docstring
"""
function save_array(Aname,A)
fname = string(Aname,".bin")
out = open(fname,"w"); write(out,A); close(out)
end
"""
Some docstring
"""
function load_array(Aname,A)
fname = string(Aname,".bin")
fid=open(fname,"r"); read!(fid,A); close(fid)
endAdd to the bin_io_script.jl a main() function that will:
generate a 3x3 array A of random numbers
initialise a second array B to hold the read-in results
call the save_array function and save the random number array
call the load_array function and read the random number array in B
return B
call the main function making and plotting as following
B = main()
heatmap(B)Make the Literate-based workflow to automatically build on GitHub using GitHub Actions. For this, you need to add to the .github/workflow folder (the one containing your CI.yml for testing) the Literate.yml script which we saw in this lecture's section Documentation tools: Automating Literate.jl.
That's all! Head to the Project section in Logistics for a check-list about what you should hand in for this first project.